The figure demonstrates the embedding of an interface $partialOmega$ into a discrete grid using the signed Euclidean distance function. The distances are computed using 4 iterations of a Newton-Raphson algorithm. The image is similar to the one in my paper:
@article{chang2010tracking,
title={Tracking monotonically advancing boundaries in image sequences
using graph cuts and recursive kernel shape priors},
author={Chang, J. and Chou, T. and Brennan, K.},
journal={Medical Imaging, IEEE Transactions on},
number={99},
pages={1--1},
year={2010},
publisher={IEEE}
}
The example is based on the SWAN wave model
by Marco Miani.
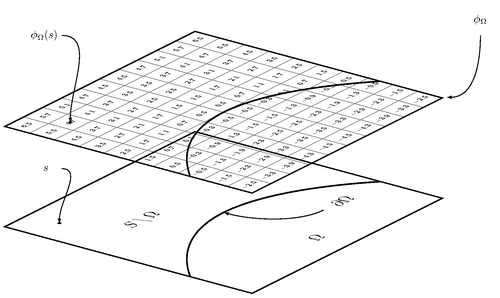
Edit and compile if you like:
% Using signed distance functions to embed contours in discrete grids
% Author: Josh Chang
\documentclass{article}
\usepackage{tikz}
\usepackage[active,tightpage]{preview}
\PreviewEnvironment{tikzpicture}
\setlength\PreviewBorder{5pt}%
\usetikzlibrary{positioning}
\usetikzlibrary{calc}
\usetikzlibrary{arrows,shapes,backgrounds}
\begin{document}
\begin{tikzpicture}[scale=2,every node/.style={minimum size=1cm},on grid]
% slanting: production of a set of n 'laminae' to be piled up.
% N=number of grids.
\begin{scope}[
yshift=-100,every node/.append style={
yslant=0.5,xslant=-1.3},yslant=0.5,xslant=-1.3
]
% opacity to prevent graphical interference
\fill[white,fill opacity=0.9] (0,0) rectangle (4,4);
\draw[step=4mm, thin, gray] (0,0) grid (4,4); %defining grids
\draw[black,very thick] (0,0) rectangle (4,4);%marking borders
\draw [ultra thick](0,1) parabola bend (2,2) (4,1) ; % parabola curve
\coordinate (sphi) at (0.6,3.4);
\node at (sphi) [fill=black,circle,scale=0.1] {$s$};
\pgfkeys{/pgf/number format/.cd, fixed, zerofill, precision =1}
\foreach \x in {0,...,9} {
\foreach \y in {0,...,9} {
%calculate the signed distance
%
% use newton raphson for 4 iterations to compute the distance
%
\pgfmathparse{0.2+0.4*\x}
\pgfmathresult \let\xpoint\pgfmathresult;
\pgfmathparse{0.2+0.4*\y}
\pgfmathresult \global\let\ypoint\pgfmathresult;
\pgfmathparse{\xpoint}
\pgfmathresult \global\let\xx\pgfmathresult;
% Run 4 iterations of Newton-Raphson to compute distance
\foreach \iter in {1,...,4} {
\pgfmathparse{0.25*(\xx*\xx*\xx-6*\xx*\xx+4*(\xx-2)*\ypoint
+12*\xx-8*\xpoint+8)}
\pgfmathresult \let\functionderv\pgfmathresult;
\pgfmathparse{3*(\xx-2)*(\xx-2)/4+\ypoint}
\pgfmathresult \let\functiondervv\pgfmathresult;
\pgfmathparse{\xpoint-\functionderv/(\functiondervv)}
\pgfmathresult \let\xx\pgfmathresult;
}
\pgfmathparse{-\xx*\xx/4+\xx+1}
\pgfmathresult \global\let\yy\pgfmathresult;
\pgfmathsetmacro{\dd}{sqrt((\xpoint-\xx)* (\xpoint-\xx)
+ (\ypoint-\yy)*(\ypoint-\yy ))/.4};
\pgfmathparse{int(\yy*100)}
\pgfmathresult \let\yyy\pgfmathresult;
\pgfmathparse{int(\ypoint*100)}
\pgfmathresult \let\yypoint\pgfmathresult;
\ifnum \yyy > \yypoint { %% Signed distance
\pgfmathparse{-\dd} \pgfmathresult \global\let\dd\pgfmathresult;
}
\fi
\node[scale=0.7,thick] at (\xpoint,\ypoint)
{$\mathbf{\pgfmathprintnumber{\dd}}$};
}
}
\end{scope}
\begin{scope}[
yshift=-160,every node/.append style={
yslant=0.5,xslant=-1.3},yslant=0.5,xslant=-1.3
]
%marking border
\draw[black,very thick] (0,0) rectangle (4,4);
%draw bottom parabola
\draw [ultra thick](0,1) parabola bend (2,2) (4,1) ;
\draw[-latex,thick](2.8,1)node[right,scale=1.5]{$\partial\Omega$}
to[out=180,in=270] (2,1.99);
\node at (2,0.5) [scale=1.5] {$\Omega$};
\node at (1.2,2.7) [scale=1.5] {$S\setminus\Omega$};
\coordinate (s) at (0.5,3.5);
\node at (s) [fill=black,circle,scale=0.1] {$s$};
\end{scope} %end of drawing grids
% signed distance
\draw[-latex,thick](4.8,-.2)node[above,scale=1.3]{$\phi_\Omega$}
to[out=-90,in=0] (4.1,-1.5);
% s
\draw[-latex,thick](-4,-.2)node[left,scale=1.3]{$\phi_\Omega(s)$}
to[out=0,in=90] (sphi);
%s
\draw[-latex,thick](-4,-3)node[left,scale=1.3]{$s$}
to[out=0,in=90] (s);
\end{tikzpicture}
\end{document}
Click to download: contours-grids.tex • contours-grids.pdf
Open in Overleaf: contours-grids.tex